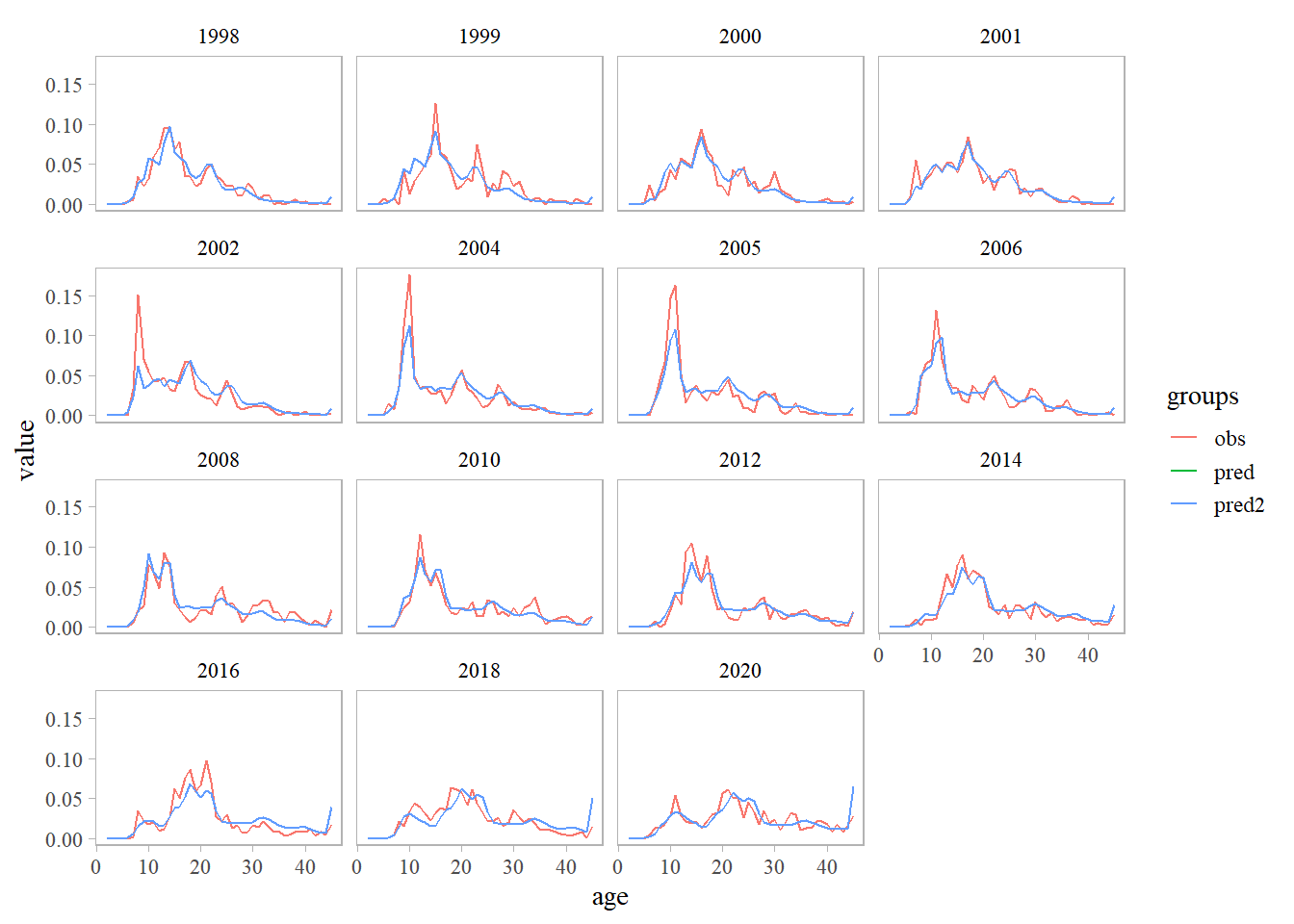
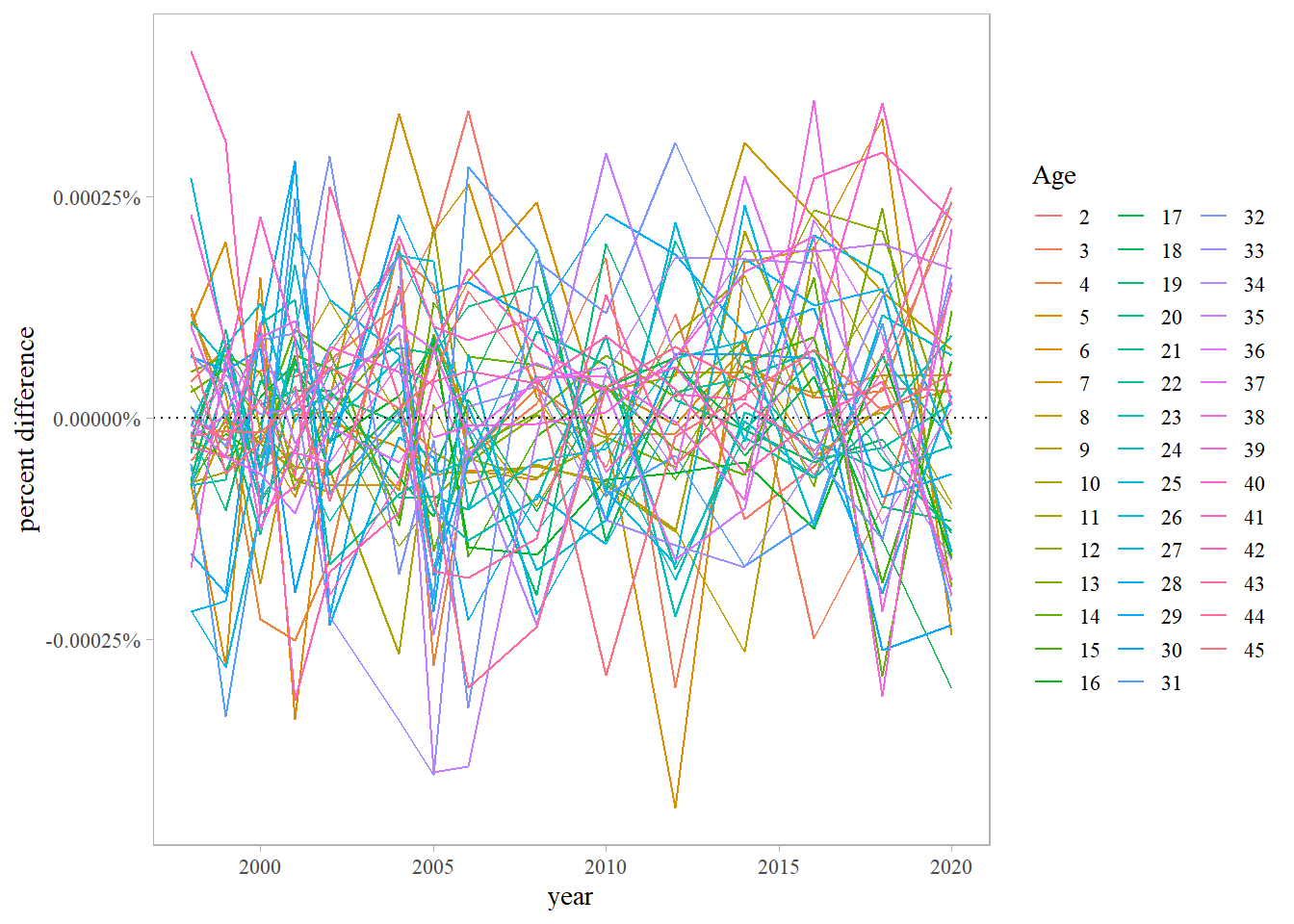
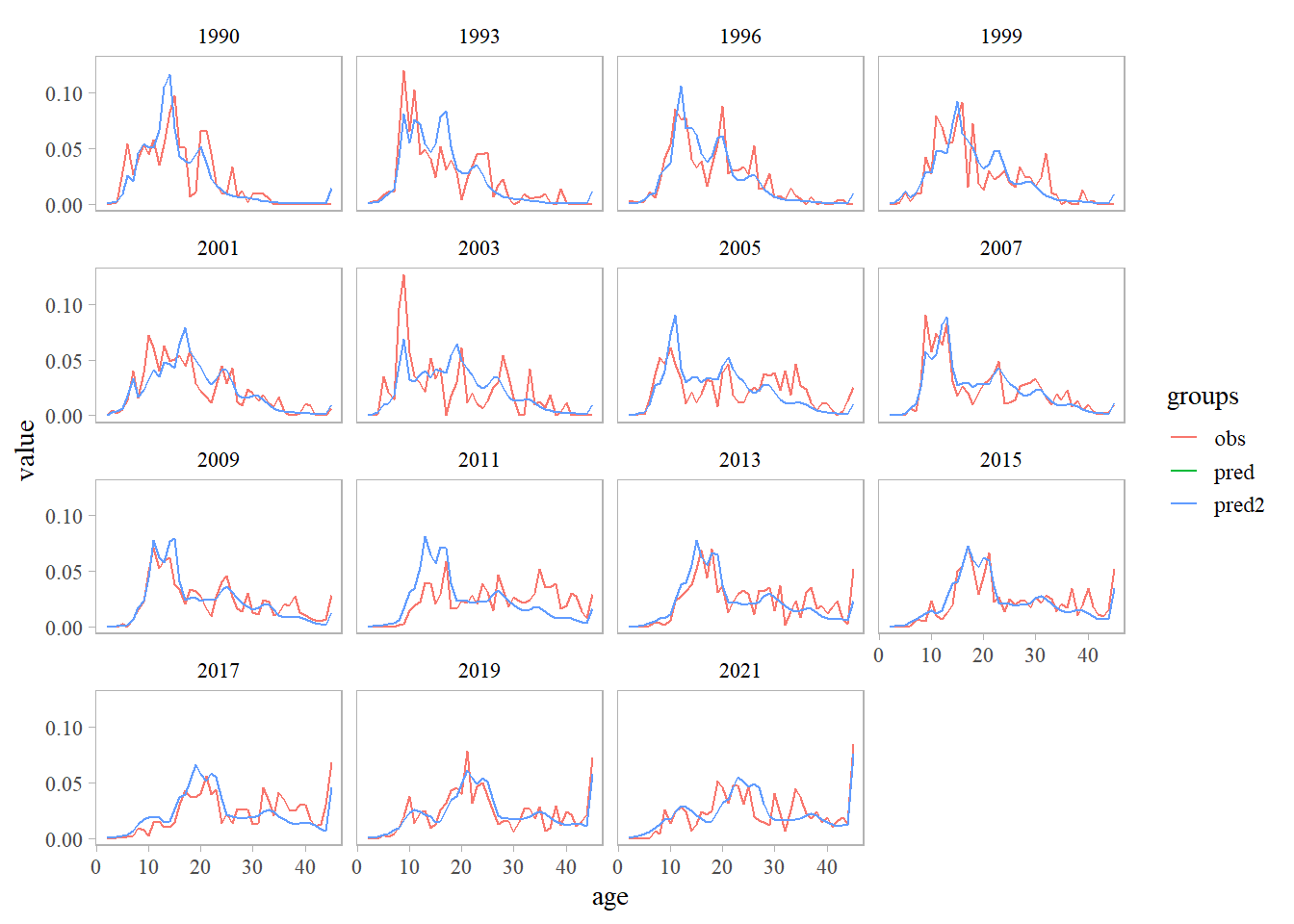
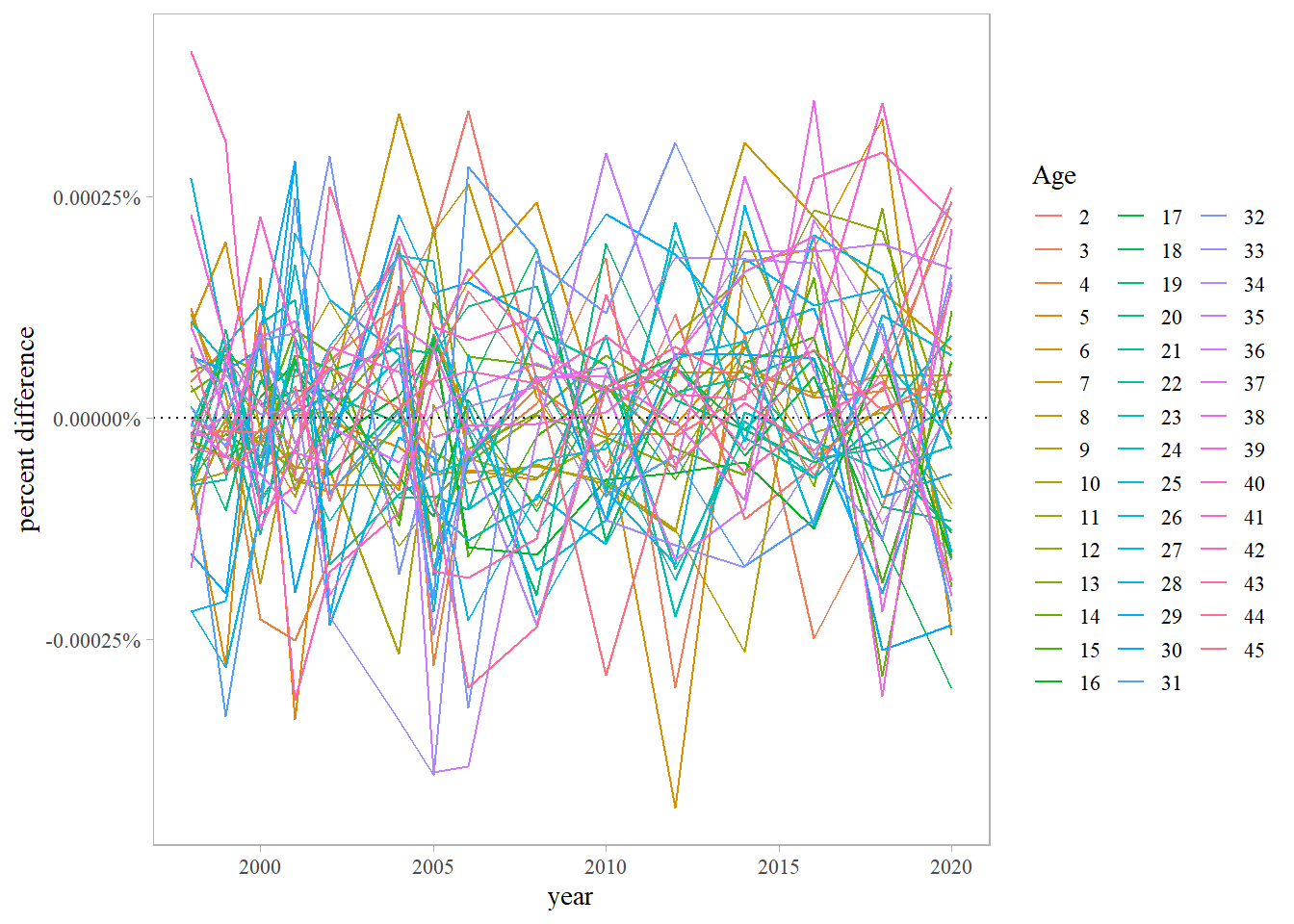
# retrieve data/results from ADMB & RT<B models
data <- source(here::here('data', 'nork_data_2022.r'))
rtmb <- readRDS(here::here('data', 'rtmb.RData'))
rtmb_proj <- readRDS(here::here('data', 'rtmb_proj.RData'))
catch <- read.csv(here::here('data', 'catch.csv'))
srv <- read.csv(here::here('data', 'survey.csv'))
fac <- read.csv(here::here('data', 'fac.csv'))
sac <- read.csv(here::here('data', 'sac.csv'))
fsc <- read.csv(here::here('data', 'fsc.csv'))
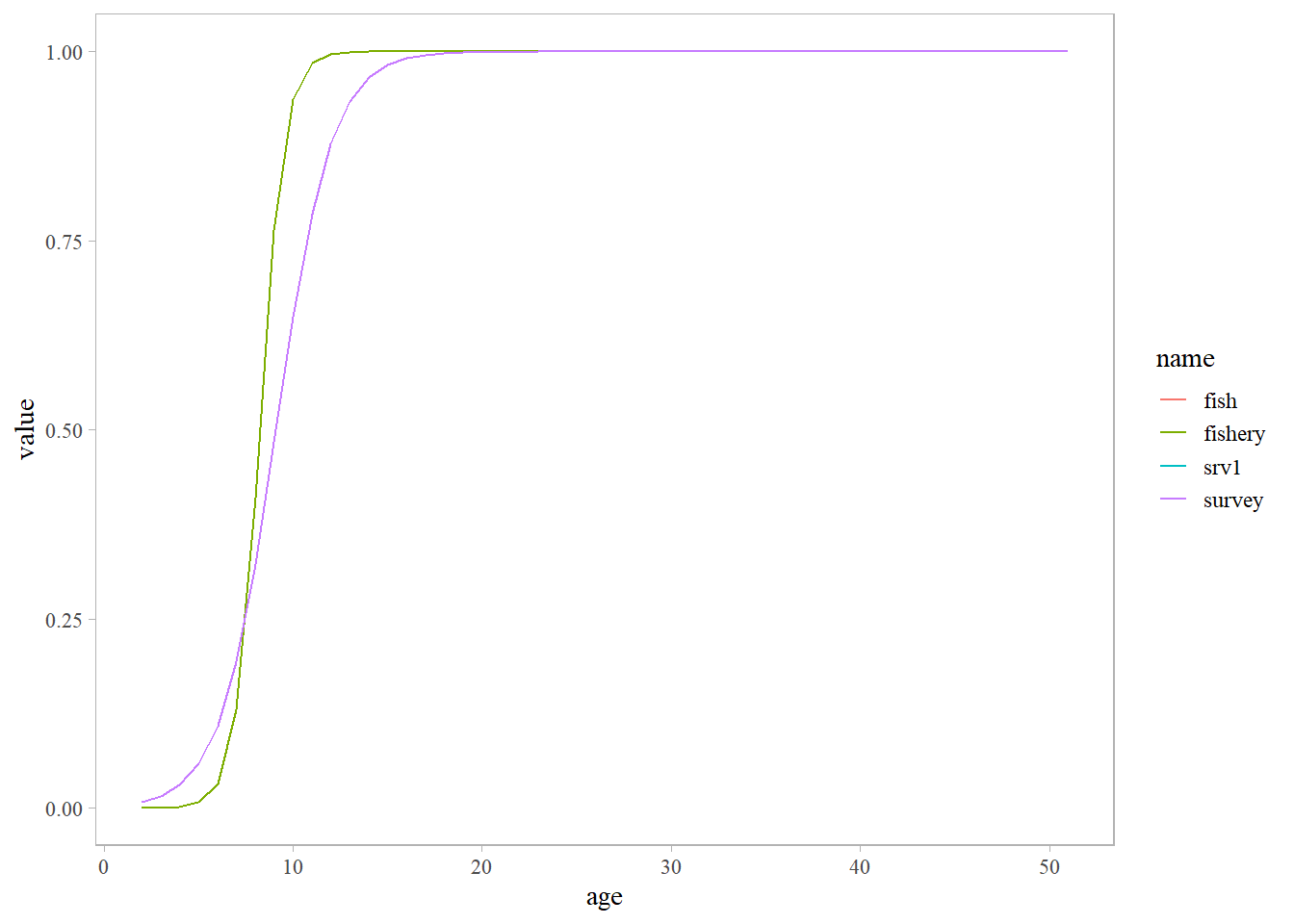
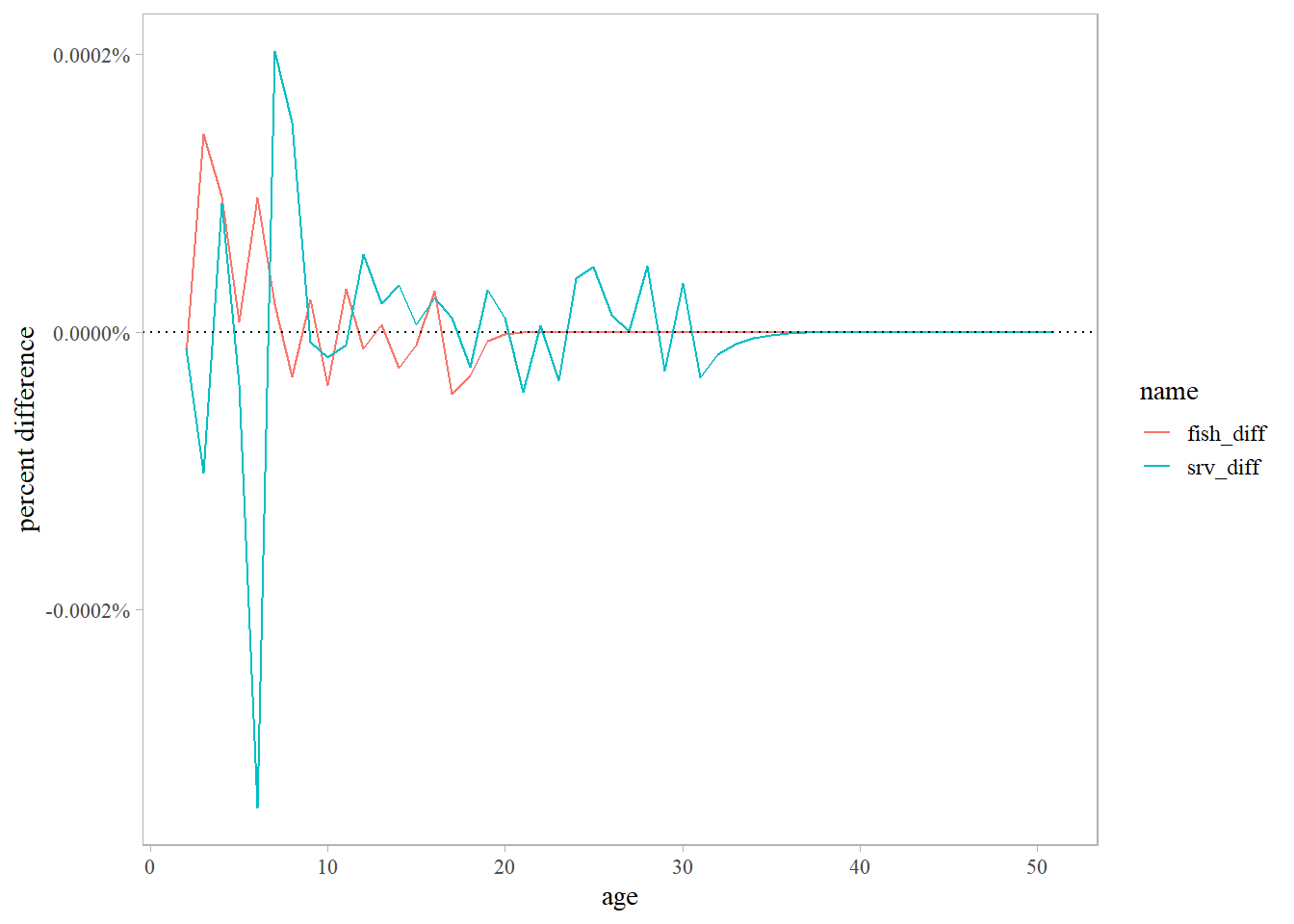
slx <- read.csv(here::here('data', 'selex.csv'))
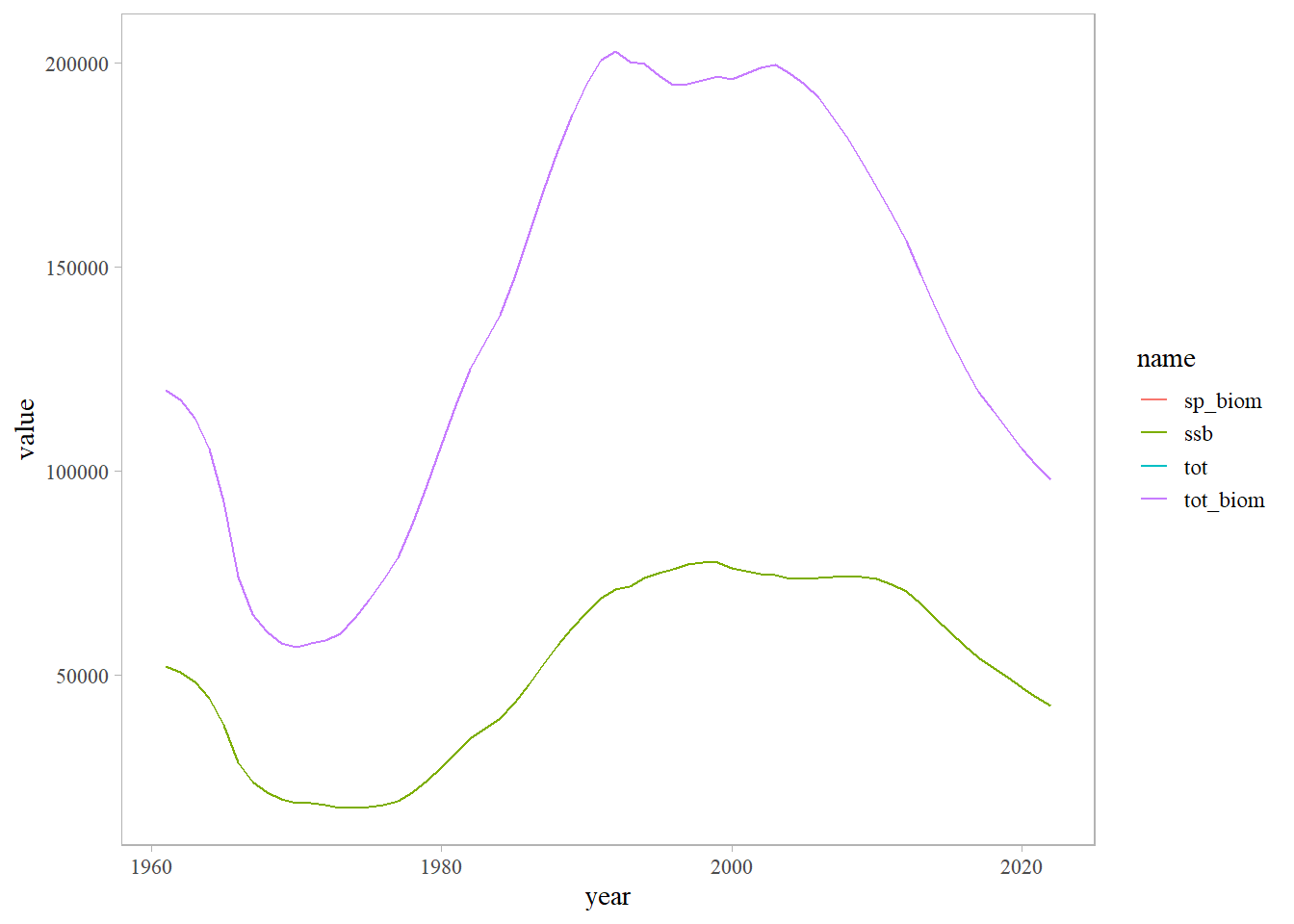
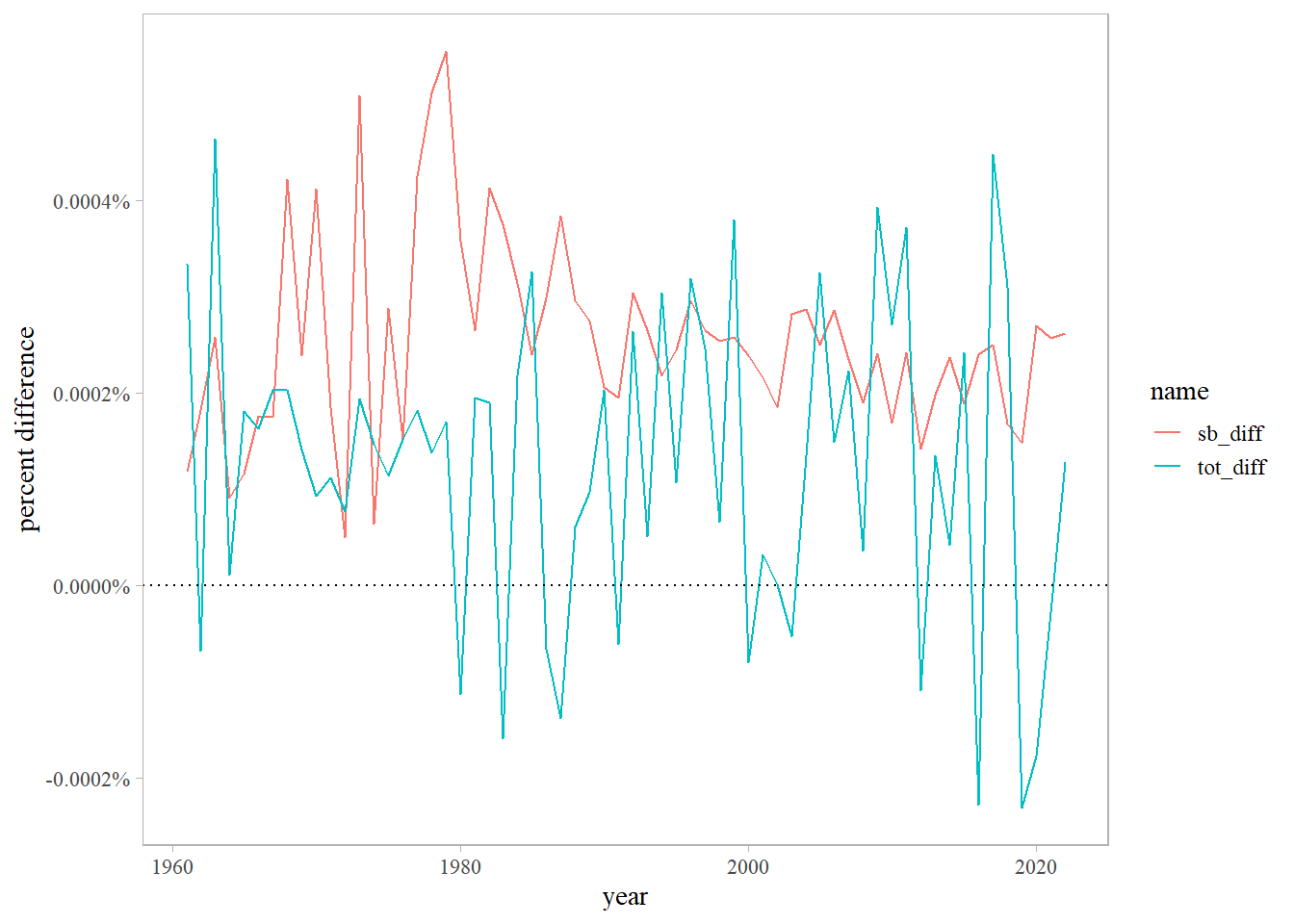
bio <- read.csv(here::here('data', 'bio_rec_f.csv'))
n_proj <- read.csv(here::here('data', 'n_proj.csv'))
b40 <- read.csv(here::here('data', 'b35_b40_yld.csv'))